Overview
RNA helicases are molecular machines that use the energy of ATP hydrolysis to unwind RNA secondary structures and remodel RNA-protein complexes. All organisms rely on these proteins to conduct RNA biogenesis processing, translation, and decay. In a human cell, there are over 60 RNA helicases and tens of thousands of RNA species. Our research addresses a fundamental question: how do distinct RNA helicases, all based on a highly conserved protein architecture, manage to bind and act on the right RNAs at the right times?
We have established methods to study the function of RNA helicases in gene expression regulation that span from in vitro helicase assays to genome-wide investigations of RNA abundance by RNA-sequencing. Our group is currently leveraging these tools to study how the RNA helicase UPF1 selects RNAs for nonsense-mediated decay (NMD).
UPF1 and NMD
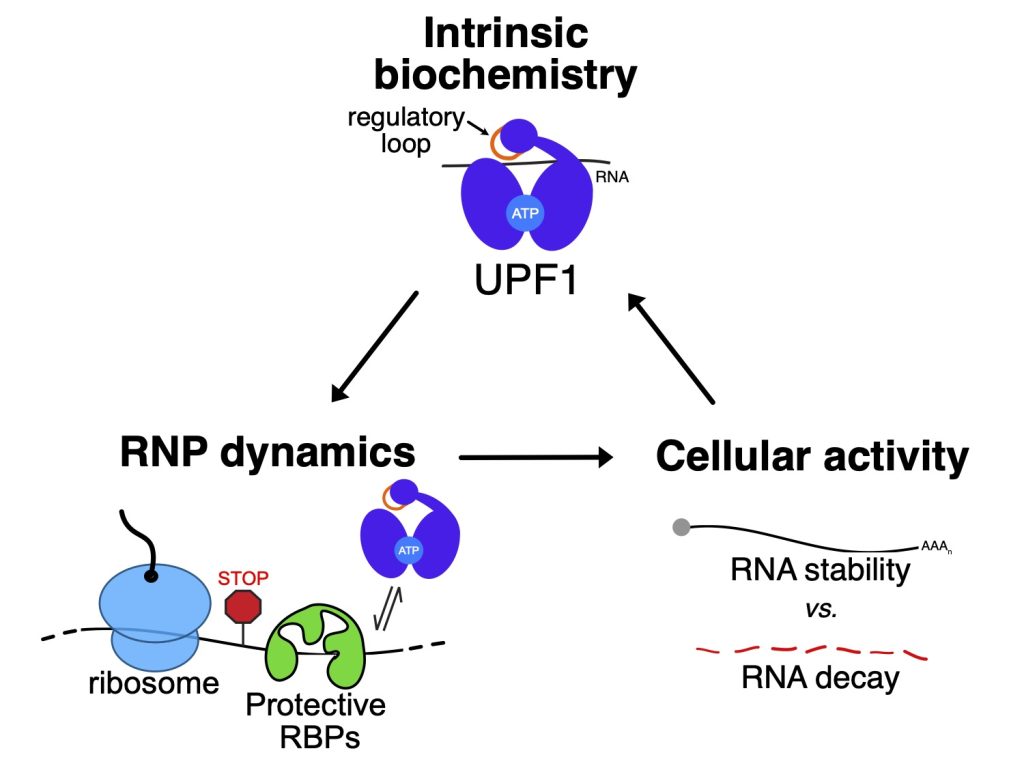
UPF1 is the central coordinator of NMD, a pathway responsible for degrading faulty protein-coding RNAs harboring premature translation termination codons as well as 5-10% of normal RNAs. Like most RNA helicases, UPF1 engages in non-specific interactions with wide variety of RNAs. However, it only commits some of them to NMD. As a postdoctoral fellow, Professor Fritz discovered that the helicase core of UPF1 uses a novel feature – a regulatory loop – to modulate which RNAs it selects for NMD. What is not known, but critical to determine, is the mechanism by which the regulatory loop controls UPF1 function in NMD target selection. In the absence of such knowledge, the development of interventions that can modulate the fate of specific RNAs for NMD will remain a challenge.
Significance
An estimated one third of human genetic diseases are affected by NMD, yet there are no FDA-approved strategies to modulate this RNA decay pathway. This unmet need is due to our poor understanding of the basic mechanisms by which RNAs are selected for NMD and the step-by-step process by which substrates are decayed. Leveraging biochemical, genetic, cellular, and molecular techniques, we aim to define the mechanism by which the UPF1 regulatory loop controls NMD target selection.
Current projects
Defining the mechanism of the regulatory loop in modulating UPF1 helicase activity.
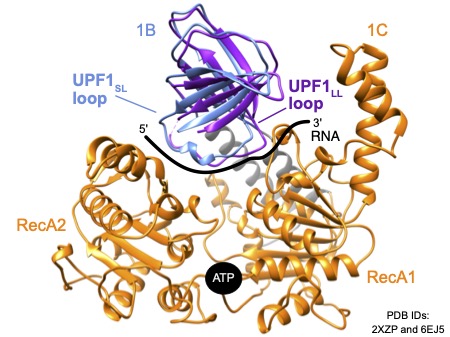
The UPF1 regulatory loop is a conserved 11 amino acid structural element within the helicase core. The loop intrudes on the RNA binding surface, causing UPF1 to release RNA upon ATP hydrolysis. Interestingly, mammals express two UPF1 isoforms that differ only in the length of the regulatory loop: a UPF1 “short loop” isoform (UPF1SL), which harbors the 11 amino acid regulatory loop, and a UPF1 “long loop” isoform (UPF1LL) with an extended regulatory loop of 22 amino acids. The longer UPF1 regulatory loop isoform has increased catalytic activity and a higher affinity for RNA in the presence of ATP than the shorter UPF1 regulatory loop isoform. In her postdoctoral work, Professor Fritz discovered that the distinct biochemical properties of the longer UPF1 regulatory loop results in differential NMD substrate specificity in human cells.
An outstanding question is how UPF1 regulatory loop sequences and structures contribute to RNA decay specificity. By studying a library of regulatory loop variants and leveraging established in vitro helicase assays, this project aims to define how regulatory loop sequence, length, and structure affect the biochemical properties of UPF1.
Understanding the function of the UPF1 regulatory loop in controlling NMD target selection.
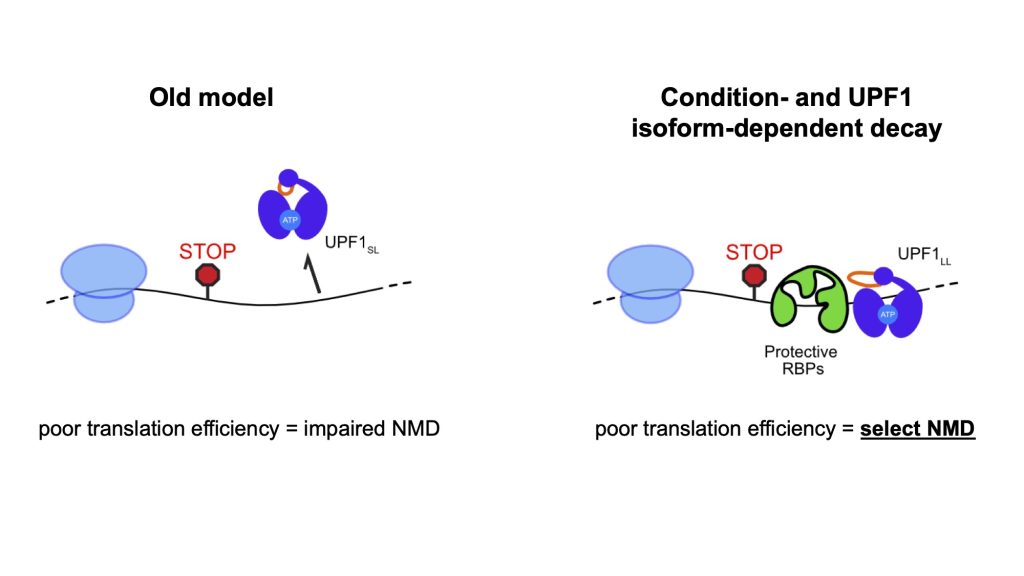
Susceptibility of a RNA to NMD can be modulated by the RNA-binding proteins PTBP1 and hnRNP L, which associate with specific transcripts and shield them from decay. As a postdoctoral fellow, Professor Fritz discovered that PTBP1 exploits the UPF1 regulatory loop to promote the ATPase-dependent dissociation of UPF1 from potential NMD substrates. She further identified that the long UPF1 regulatory loop isoform can overcome this protective mechanism to bind and down-regulate RNAs normally shielded from NMD. Unexpectedly, she discovered that the long UPF1 regulatory loop isoform can also support induction of NMD on new populations of substrate RNAs in response to cellular stress and impaired translation efficiency.
An outstanding question is how the UPF1 regulatory loop can confer differential NMD substrate specificity. By determining the effects of UPF1 regulatory loop mutants on NMD target selection using established human cell lines and RNA-sequencing, this project aims to understand how the biochemical activity of the regulatory loop enables UPF1 to control RNA decay target selection in response to negative regulatory factors and changing cellular states.
Characterizing the role of the regulatory loop in modulating activation of UPF1 by UPF2.
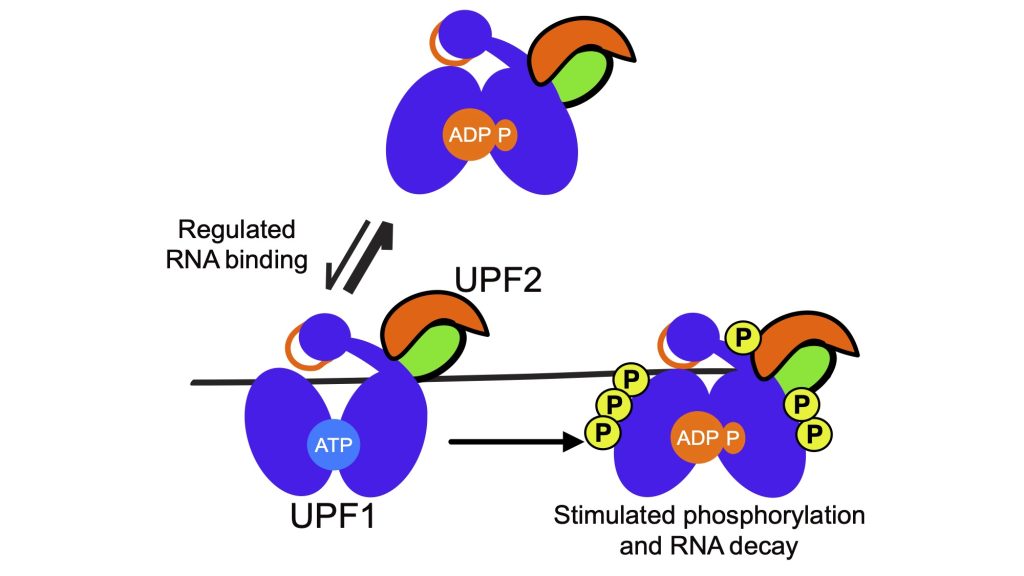
UPF2 is a cofactor known to activate UPF1 helicase activity and promote its phosphorylation in cells. These stimulatory effects of UPF2 on UPF1 function are critical in the commitment of a RNA to NMD. However, recent studies have indicated that this relationship is more complex, with UPF2 acting to regulate initial binding of UPF1 to a substrate then promoting its dissociation. These data indicate that enhancement of UPF1 helicase activity by UPF2 may also function to regulate initial NMD target selection, with enhanced dissociation of UPF1 from a potential substrate by UPF2 resulting in RNA stability rather than decay.
An outstanding question is the extent to which the regulatory loop can control the activation of UPF1 by UPF2 and the role of this interaction in governing NMD target specificity. Leveraging our library of regulatory loop mutants and established biochemical and cellular assays, this project aims to characterize how the intrinsic biochemical activity of the regulatory loop can control the degree to which UPF2 activates UPF1 helicase activity and influences NMD target selection in cells.
Funding
We thank the following sources of support for our work:
- Villanova University, College of Liberal Arts and Sciences, start-up fund to SEF
- Villanova University, College of Liberal Arts and Sciences, GRASP award to SEF
- Villanova University, University Summer Grant to SEF
- Villanova University, Department of Biology, BIO-SUGR award to Lillian Holzinger, Michael Blake, and Halle Roberts
- Villanova University, VURF award to Emma Thomas

